
Immunoglobulins are crucial proteins produced by the immune system to identify and bind to foreign substances, playing an essential role in shielding organisms from infections and diseases. Designing specific antibodies opens new pathways for disease treatment. With the rise of deep learning, AI-driven drug design has become possible, leading to several methods for antibody design. However, many of these approaches require additional conditions that differ from real-world scenarios, making it challenging to incorporate them into existing antibody design processes. Here, we introduce IgGM, a generative model for the de novo design of immunoglobulins with functional specificity. IgGM simultaneously generates antibody sequences and structures for a given antigen, consisting of three core components: a pre-trained language model for extracting sequence features, a feature learning module for identifying pertinent features, and a prediction module that outputs designed antibody sequences and the predicted complete antibody-antigen complex structure. IgGM effectively predicts structures and designs novel antibodies and nanobodies. This makes it highly applicable in a wide range of practical situations related to antibody and nanobody design.
For more details, please refer to the paper.
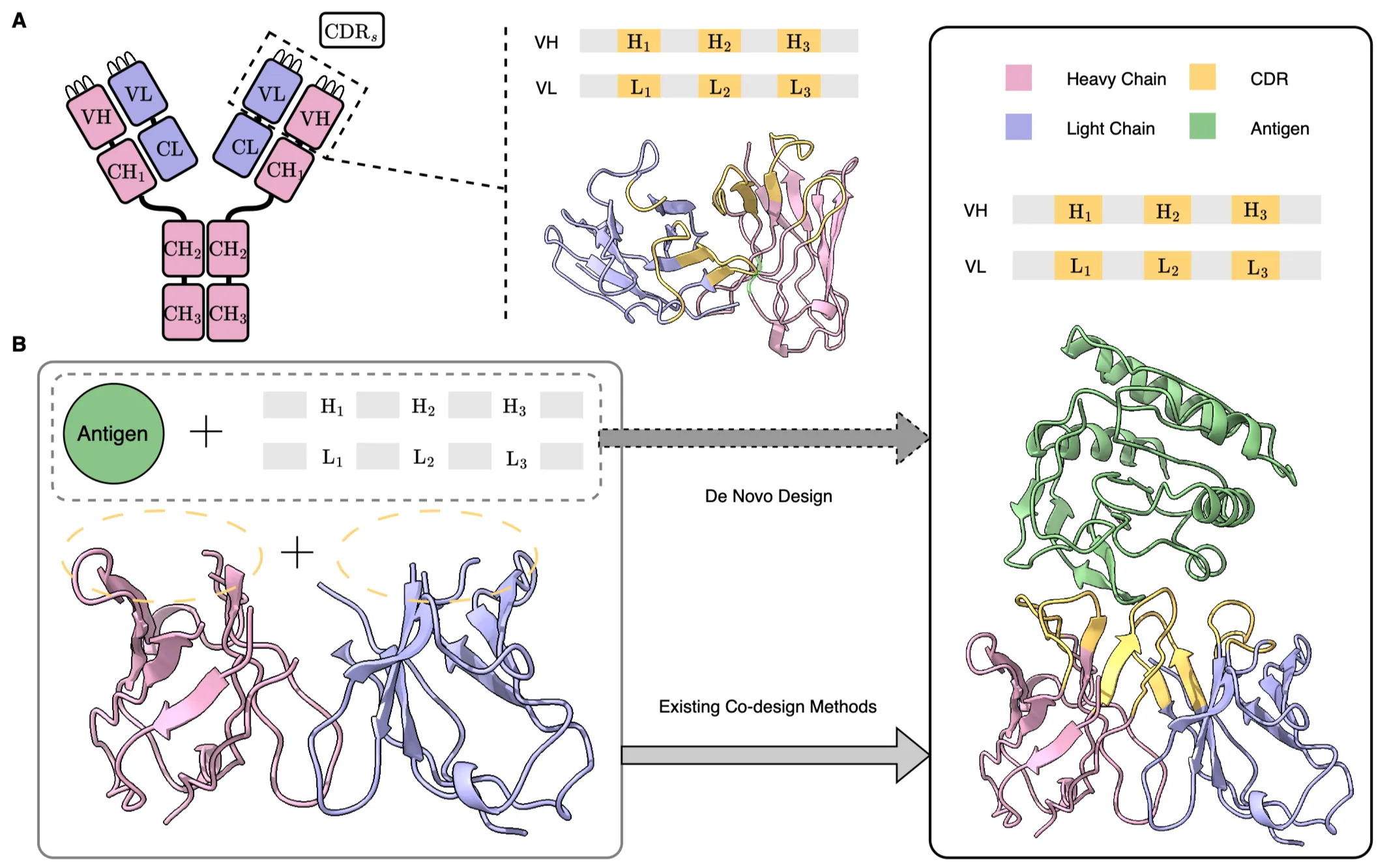
An antibody consists of a symmetric Y-shaped structure, which includes variable regions (VH, VL) and constant regions (CH, CL). In practical antibody design, the focus is on the variable regions, which comprise the framework regions (FRs) and the complementarity-determining regions (CDRs).
De novo antibody design refers to the process of creating a novel antibody that can bind to a given antigen, where the framework regions can be selected based on sequences with favorable physicochemical properties. Existing co-design methods require the simultaneous provision of both the structure and sequence of the framework regions; however, in practical antibody design, the structures are often unknown.
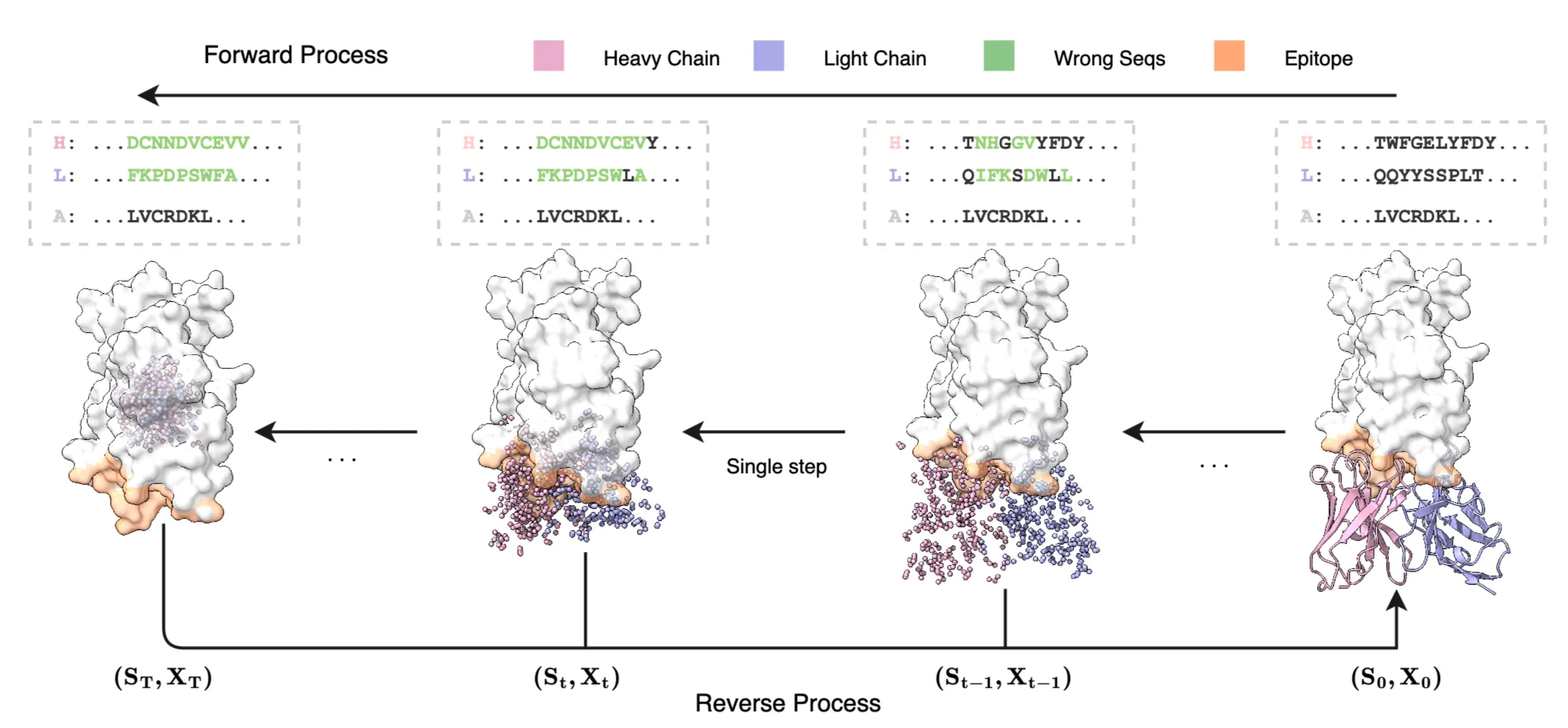
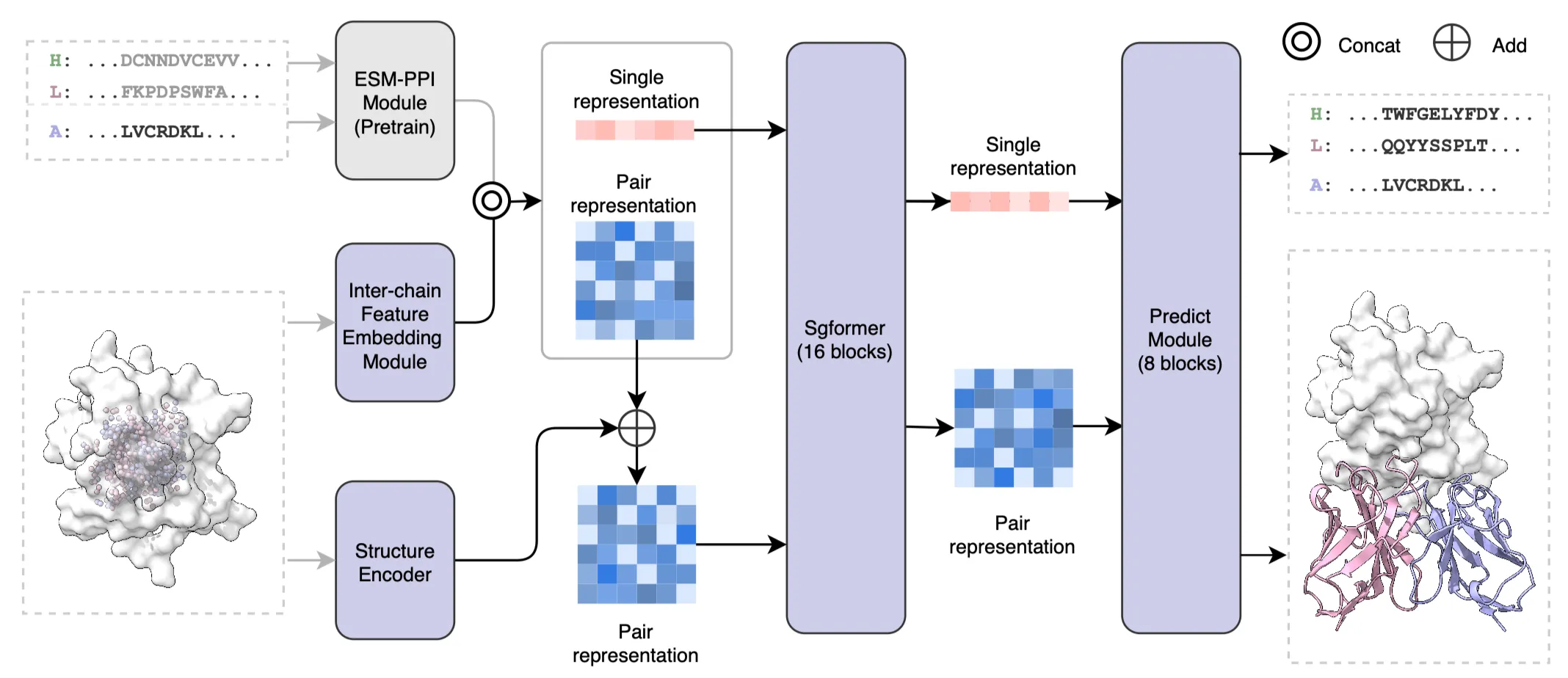
IgGM is a generative model that performs simultaneous co-design of antibody sequence and structure. IgGM employs a multi-level network architecture. It first utilizes a pre-trained protein language model to extract evolutionary features of sequences. Then, a feature encoder studies the interactions between antigens and antibodies. Finally, a prediction module outputs the structures and sequences of the antibodies. IgGM leverages the interplay between sequence and structure to generate accurate antibody designs, even when only partial sequences of the framework region are available. This capability aligns with practical application scenarios and offers new possibilities for antibody design. IgGM excels at generating the CDR regions and their structures and can dock the generated structure to the corresponding epitope. It supports multiple design scenarios and can adapt to various conditions without the need for retraining, such as predicting antigen-antibody complexes, designing the CDR H3 region of antibodies, and designing multiple CDR regions. Furthermore, it can be extended to nanobodies, which are small single-domain antibodies that exhibit strong binding affinity to antigens and high stability. The experimental results indicate that IgGM achieves superior performance in multiple design tasks, demonstrating accuracy in structure prediction tasks that is comparable to existing structure prediction methods.
.BkZ6b6De_Z2wK86m.webp)
| Method | TM-Score↑ | lDDT↑ | RMSD↓ | DockQ↑ | iRMS↓ | LRMS↓ | SR↑ | |
|---|---|---|---|---|---|---|---|---|
| IgFold†→HDock | 0.9577 | 0.9019 | 2.1715 | 0.0218 | 16.6519 | 48.1571 | 0.0000 | |
| tFold-Ag*‡ | 0.9634 | 0.9142 | 1.9489 | 0.2522 | 6.7957 | 21.0346 | 0.4068 | |
| AlphaFold 3‡ | 0.9729 | 0.9305 | 1.5063 | 0.2951 | 10.9645 | 32.4080 | 0.3684 | |
| dyMEAN†* | 0.9572 | 0.8882 | 2.2521 | 0.1005 | 8.9227 | 27.4234 | 0.0667 | |
| IgGM†* | 0.9591 | 0.8956 | 2.1997 | 0.2986 | 6.2195 | 19.4888 | 0.4667 | |
| IgGM†*(AF3) | 0.9580 | 0.8941 | 2.1422 | 0.3630 | 3.8635 | 11.2647 | 0.6667 |
| Metrics | DiffAb (IgFold) | DiffAb (AF3) | MEAN (IgFold) | MEAN (AF3) | dyMEAN | IgGM | IgGM (AF3) | |
|---|---|---|---|---|---|---|---|---|
| AAR↑ | L1 | 0.597 | 0.608 | - | - | 0.633 | 0.750 | 0.737 |
| L2 | 0.598 | 0.599 | - | - | 0.634 | 0.743 | 0.735 | |
| L3 | 0.421 | 0.424 | - | - | 0.570 | 0.635 | 0.602 | |
| H1 | 0.642 | 0.637 | - | - | 0.742 | 0.740 | 0.739 | |
| H2 | 0.363 | 0.394 | - | - | 0.627 | 0.644 | 0.639 | |
| H3 | 0.214 | 0.226 | 0.248 | 0.246 | 0.294 | 0.360 | 0.330 | |
| RMSD↓ | L1 | 0.783 | 0.749 | - | - | 0.864 | 0.589 | 0.659 |
| L2 | 0.471 | 0.466 | - | - | 0.481 | 0.378 | 0.395 | |
| L3 | 1.002 | 1.017 | - | - | 0.941 | 0.847 | 0.903 | |
| H1 | 0.650 | 0.623 | - | - | 0.633 | 0.555 | 0.590 | |
| H2 | 0.641 | 0.586 | - | - | 0.705 | 0.486 | 0.566 | |
| H3 | 2.741 | 2.646 | 2.357 | 2.300 | 2.454 | 2.131 | 2.155 | |
| Docking | DockQ↑ | 0.022 | 0.208 | 0.022 | 0.207 | 0.079 | 0.246 | 0.326 |
| iRMS↓ | 17.034 | 9.731 | 16.838 | 8.968 | 9.698 | 6.579 | 4.030 | |
| LRMS↓ | 48.163 | 27.559 | 48.104 | 27.557 | 28.764 | 19.678 | 11.229 | |
| SR↑ | 0.000 | 0.368 | 0.000 | 0.354 | 0.049 | 0.433 | 0.627 |
| Method | TM-Score↑ | lDDT↑ | RMSD↓ | DockQ↑ | iRMS↓ | LRMS↓ | SR↑ | |
|---|---|---|---|---|---|---|---|---|
| tFold-Ag | 0.9344 | 0.9303 | 1.6722 | 0.2881 | 6.3490 | 15.0810 | 0.4296 | |
| AlphaFold 3 | 0.9519 | 0.9286 | 1.1885 | 0.2867 | 11.2194 | 32.6760 | 0.3885 | |
| IgGM | 0.9318 | 0.8931 | 1.9925 | 0.2907 | 7.9879 | 22.0168 | 0.4400 |
| Method | CDR1↑ | CDR2↑ | CDR3↑ | RMSD↓ | DockQ↑ | iRMS↓ | LRMS↓ | SR↑ |
|---|---|---|---|---|---|---|---|---|
| DiffAb (AF3) | 0.533 | 0.291 | 0.156 | 2.274 | 0.211 | 13.265 | 35.805 | 0.346 |
| IgGM | 0.565 | 0.330 | 0.183 | 1.980 | 0.267 | 6.927 | 14.966 | 0.415 |
@inproceedings{
wang2025iggm,
title={Ig{GM}: A Generative Model for Functional Antibody and Nanobody Design},
author={Rubo Wang and Fandi Wu and Xingyu Gao and Jiaxiang Wu and Peilin Zhao and Jianhua Yao},
booktitle={The Thirteenth International Conference on Learning Representations},
year={2025},
url={https://openreview.net/forum?id=zmmfsJpYcq}
}